Fresh from the digital dissection table
To further verify the correct processing of my Cubical Marching Squares-implementation (aka: CMS) – and because I grew fed up looking at my 3D perlin-noise – I have added support for loading raw CT/MR data. There are two freely available datasets from Stanford, hosted at the Stanford Volume data archive.
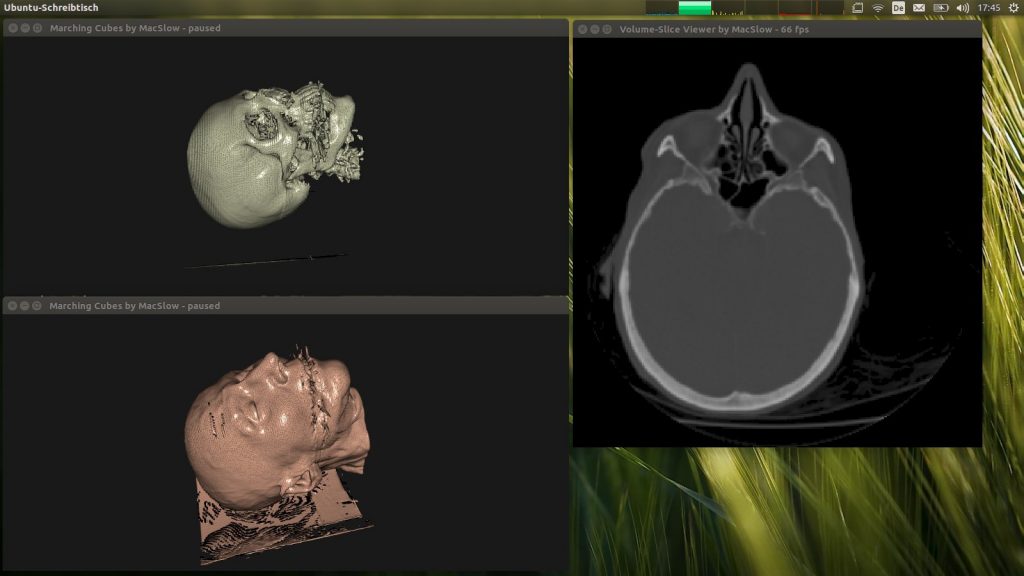
Here is my CMS-library at work on the CThead volume data-set…
A handy side-effect is that I have also implemented a viewer for these raw (16 bit, unsigned short) volume data-sets, which allows to easily scrub through all slices via the mouse-wheel. The quality of the two provided sets is not the best – as far as I can tell being a medical layman – obvious in the reflection streaks in the CThead set around the teeth (due to seals) and the overall noise in the MRbrain set. In the above screenshot you can see the “spikes” around the mouth of the patient. Those are a result of the data-acquisition artifacts.
The data-set’s resolution is also on the low side. 256x256x113 for the CThead set and 256x256x109 for the MRbrain set. This fact led to an optimization I initially planned to do a bit later… sample-value interpolation.
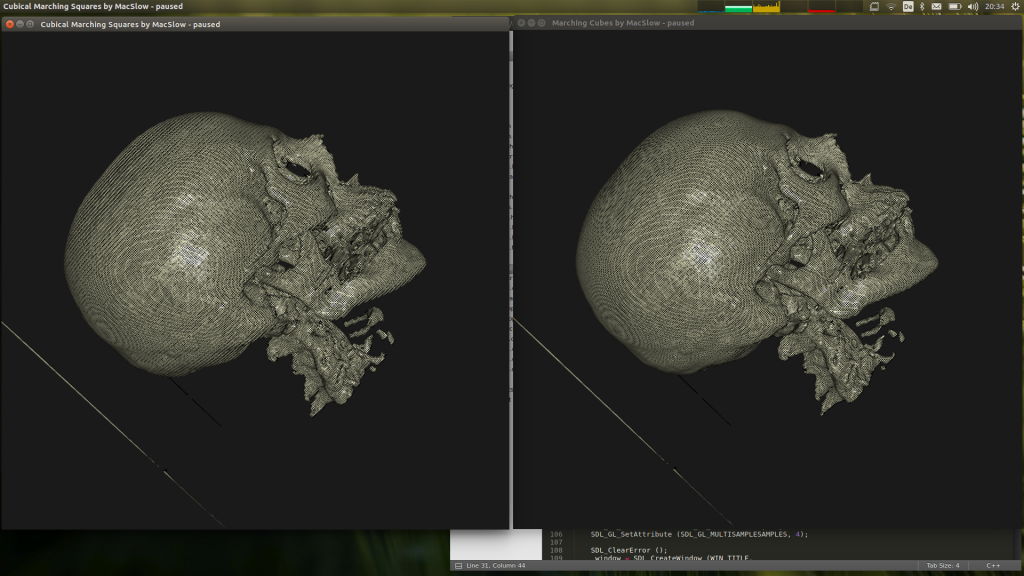
Here is a before/after screenshot showing off the supported interpolation…
Guess which is which 🙂 This interpolation counters the non-equidistant nature of the data-set lattices in question, which is roughly 1:1:2.
There are other more professional medical volume-data sets, like the male and female sets from the Visible Human Project, but these sets cannot be easily downloaded, like the ones from Stanford. By the way, Stanford only hosts the CThead and MRbrain sets. They were actually recorded by the North Carolina Memorial Hospital.
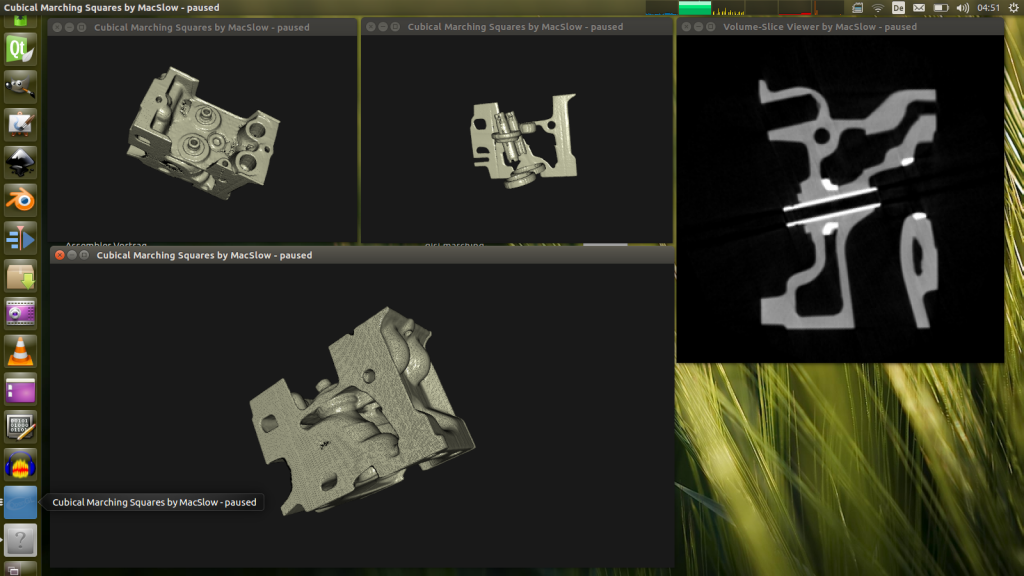
Since volume-data is volume-data, it really makes no difference what is being displayed. Non-medical data-sets work just as easily. Here is a rendition of a well-known engine-part data-set…